Engineering molecules for cellular biology
Harnessing synthetic chemistry and directed evolution methods, we aim to develop next-generation chemical probes and genetically-encoded markers for modern light microscopy and for the discovery of unknown molecular interactions. We also explore their potential as diagnostic markers and therapeutic agents.
Research Highlights
Super-resolution and single-molecule imaging of RNA, DNA and proteins
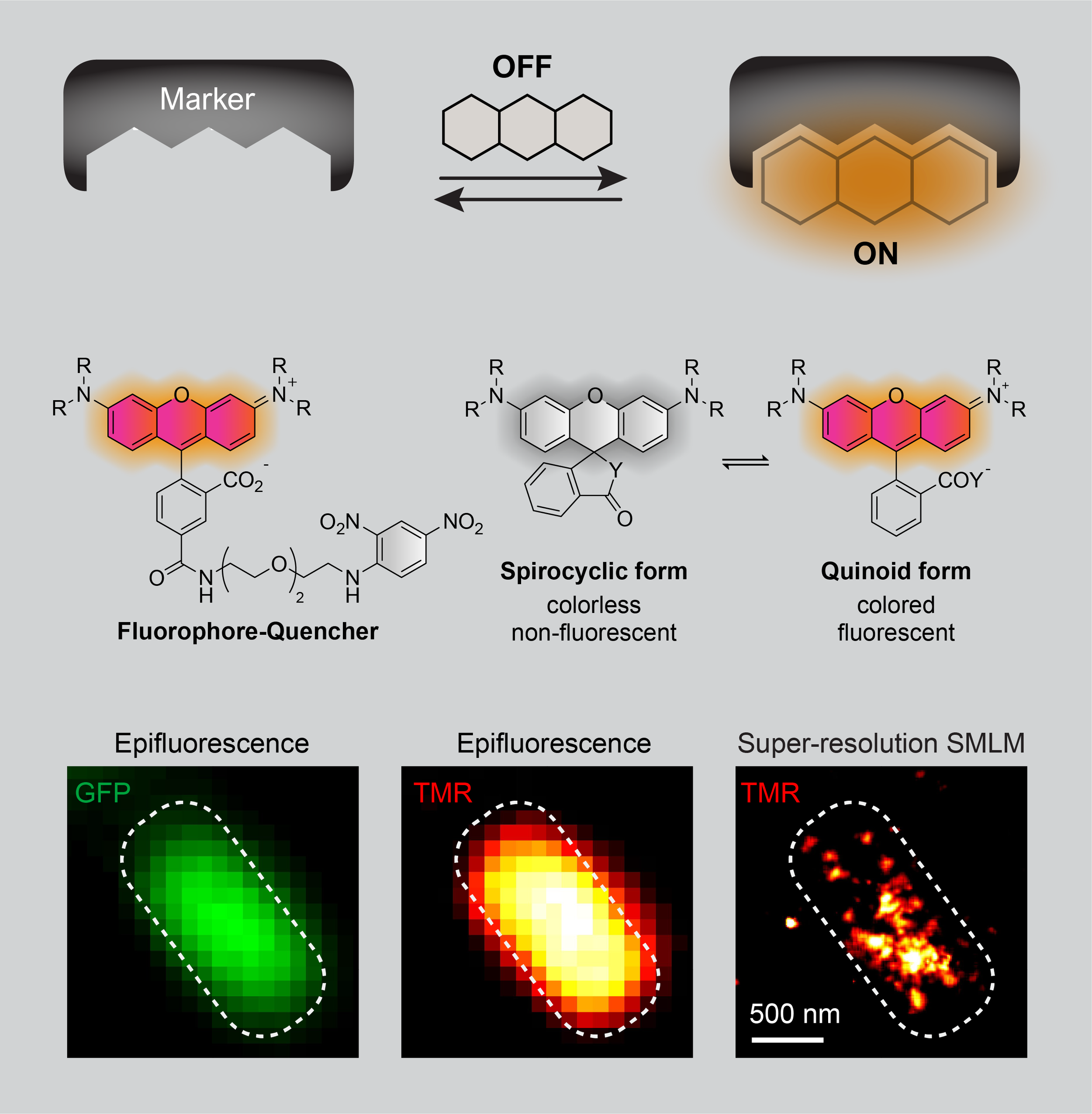
There is a great need for robust and versatile toolbox enabling super-resolution imaging of variable biomolecules with high speed and localization precision in living systems. In order to image a diverse set of biomolecules including proteins, RNA and DNA with super-resolution microscopy, we aim to generate genetically-encoded fluorescent markers that light up upon binding to membrane-permeable, non-fluorescent small molecule dyes. These tailor-made dyes will only fluoresce upon binding to their cognate peptide or nucleic acid partners and allow for no-wash fluorescence imaging. Furthermore, binding and unbinding events will result in intermittent fluorescence emission, i.e. “blinking”. This feature will enable imaging of any biomolecule of interest by single-molecule localization microscopy (SMLM) and by STED (Stimulated Emission Depletion) microscopy.
We will use directed evolution methods such as mRNA and yeast display for peptide markers, SELEX (Systematic Evolution of Ligands by EXponential Enrichment) for RNA and DNA markers, and high-throughput screening techniques such as FACS (Fluorescence Activated Cell Sorting) to select for the best light-up marker.
Multi-omics approach to decipher molecular interactions
Interactions between biomolecules are key to cellular homeostasis and their perturbation can lead to cellular malfunction and disease. Thus, new techniques for discovering and characterizing these interactions in vivo are in high demand. Although enzymatic proximity labeling methods have been broadly applied to investigate subcellular large architectures, technologies enabling microenvironment mapping with high precision are still lacking. Therefore, we aim to create novel technologies to investigate protein-protein, RNA-protein, RNA-RNA and DNA-protein interactions including weak and transient ones. We will expand known interactomes, enabling discovery of new functions of RNAs and proteins. To this end, we will use proteomics and next-generation sequencing techniques.
Aptamers for in vivo imaging and targeted photodynamic therapy
Nucleic acid aptamers can be used to generate excellent tumor tracers because they i) are small; ii) can bind target proteins selectively with high sensitivity; iii) can be rapidly distributed through tissues; iv) can be quickly (several hours) cleared from the organism unless they are bound to tumor tissue. Our goal is to generate tumor-targeting light-up markers emitting in the near-IR region for deep-tissue penetration to visualize cancer cells. These aptamers will contain modified nucleotides to render them more stable against nucleases and less immunogenic. We will also investigate approaches to enhance a cancer marker with targeted therapeutic capability by employing photodynamic therapy.
Selected Publications
* corresponding author(s)
1. Englert D, Burger E, Grün F, Verma M S, Lackner J, Lampe M, Bühler B, Schokolowski J, Nienhaus G U*, Jäschke A* and Sunbul M*
Fast-exchanging spirocyclic rhodamine probes for aptamer-based super-resolution RNA imaging
Nature Communications, 2023, 14, 3879 [Article]
2. Bühler B, Schokolowski J, Benderoth A, Englert D, Grün F, Jäschke A* and Sunbul M*
Avidity-based bright and photostable light-up aptamers for single-molecule mRNA imaging
Nature Chemical Biology, 2023, 19, 478 [Article]
3. Sunbul M*, Lackner J, Martin A, Englert D, Hacene B, Grün F, Nienhaus K, Nienhaus G U* and Jäschke A*
Super-resolution RNA imaging using a rhodamine-binding aptamer with fast exchange kinetics
Nature Biotechnology, 2021, 39, 686 [Article]
4. Zhang J, Wang L, Jäschke A* and Sunbul M*
A color-shifting near-infrared fluorescent aptamer-fluorophore module for live-cell RNA imaging
Angewandte Chemie International Edition, 2021, 60, 21441 [Article]
5. Wirth R, Gao P, Nienhaus G U, Sunbul M* and Jäschke A*
SiRA: a silicon rhodamine-binding aptamer for live-cell super-resolution RNA imaging
Journal of the American Chemical Society, 2019, 141, 7562 [Article]